RNA Biology Research in the Pasquinelli Lab
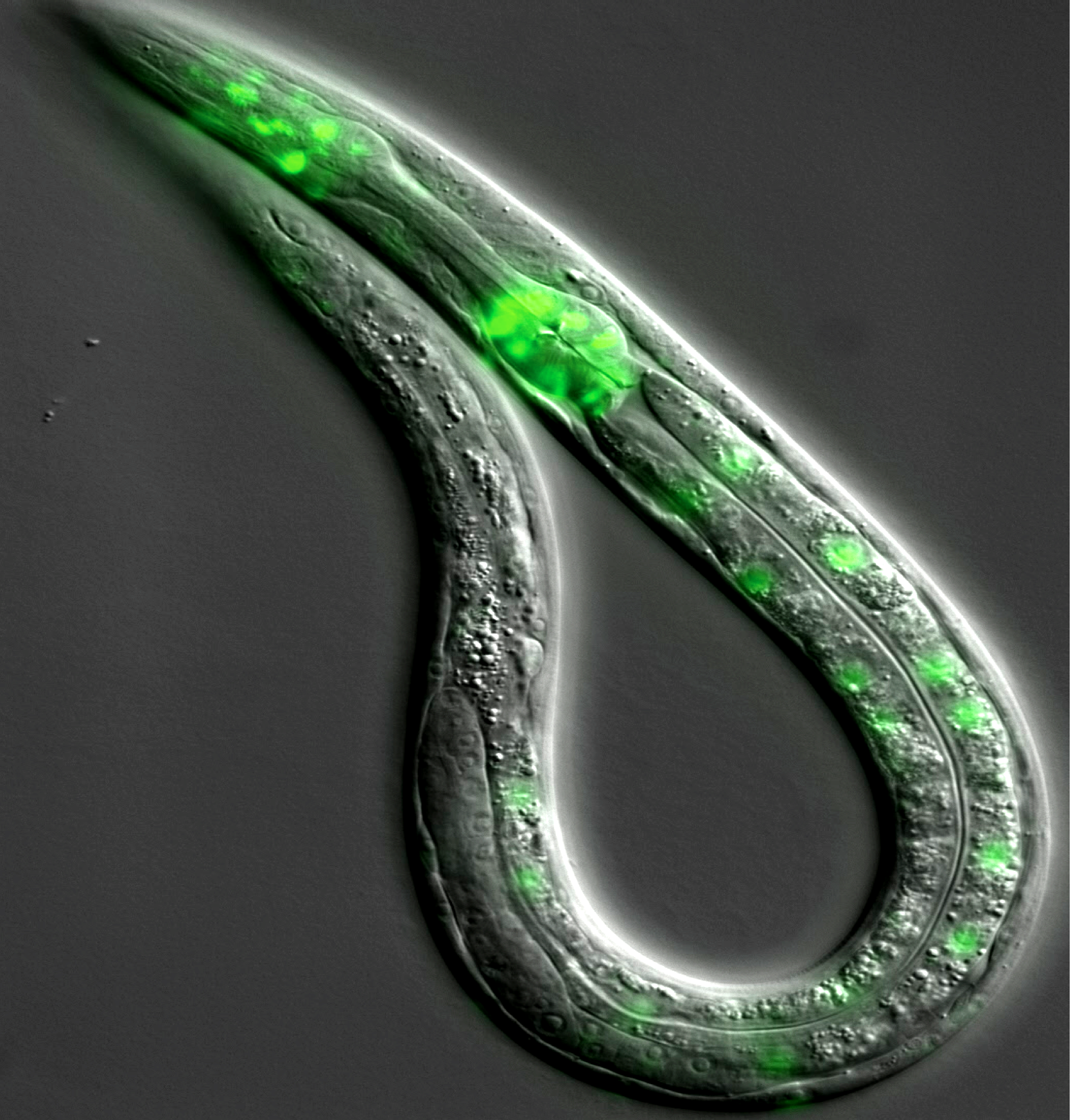
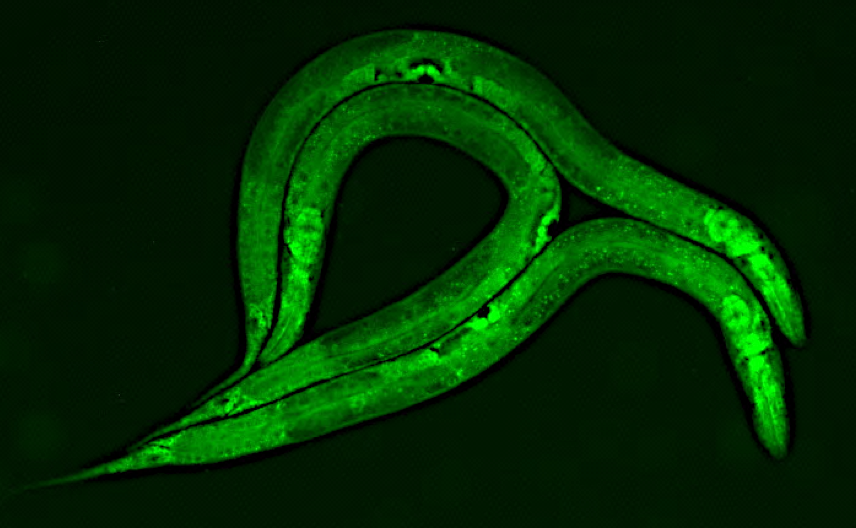
The Pasquinelli Lab is broadly interested in RNA-mediated mechanisms of gene regulation. We aim to understand how miRNAs and other non-coding regulatory RNAs are expressed and function. Much of our work has focused on miRNA biogenesis and target regulation in nematode worms called C. elegans. These animals are an ideal experimental model amenable to a wide range of tools and methods for exploring how regulatory RNA pathways function in a living organism. While some miRNAs are essential for normal development in C. elegans, others are important in particular contexts such as stress or aging. The miRNAs and pathways we study are conserved in humans and a better understanding of how they work in an endogenous context will shed light on their roles in human health and disease and contribute to the development of RNA-based therapeutics.
Mechanisms of miRNA Expression & Function
As suggested by their name, miRNAs are tiny, approximately 22 nucleotide, regulatory RNAs. However, miRNA genes typically encode long primary transcripts that undergo multiple processing steps to generate a functional miRNA. Many miRNA genes are expressed at precise times in development and in specific tissues. To understand how these temporal and spatial expression patterns are achieved, we study the transcriptional and processing events that cooperate to produce specific miRNAs at the right time and in the right place.
MiRNAs regulate specific genes by partially base-pairing to complementary sequences in the messenger RNAs (mRNAs) of protein-coding genes. The human genome contains over 1,000 different miRNA genes, each of which may directly regulate hundreds of protein coding genes. To help elucidate how miRNAs find and regulate targets with limited sequence complementarity, we use a combination of molecular methods, genomics and bioinformatics to identify potential targets. Once a target is bound by a miRNA, it undergoes mRNA degradation or translational repression, but the molecular mechanisms behind these inhibitory strategies are not entirely understood. By studying defined miRNA and target pathways in C. elegans, we aim to figure out how miRNAs control gene expression in the endogenous context.
A daunting challenge in the miRNA field is to establish biological functions for the thousands of miRNAs that have been discovered across species. While some miRNAs have proven to play essential roles in animal development, behavior, and viability, deficiencies in many other miRNAs appear to have no obvious consequences in worm or mouse knockout models. One explanation for these findings is that some miRNAs might only be needed in particular contexts, such as stress, that organisms regularly experience in the wild, but not in the laboratory. We use a combination of genetics, genomics, molecular biology and biochemistry to understand how the miRNA pathway responds to and regulates gene expression changes needed for an organism to survive environmental assaults, like high heat.
Another context where the miRNA pathway plays a key role is during organismal aging. MiRNAs depend on Argonaute (AGO) proteins to bind and regulate the expression of target genes at the post-transcriptional level. Surprisingly, we found that two AGOs play opposing roles in aging C. elegans. Loss of AGO Like Gene 1 (ALG-1) results in a shortened lifespan, while loss of ALG-2 leads to an extended lifespan. To understand the basis for these opposing longevity functions, we aim to identify the miRNAs, targets, tissues, and aging pathways involved.
The long string of adenosines added to the 3’ ends of most eukaryotic mRNAs, called the poly(A) tail, serves a wide range of functions – promoting nuclear export, protecting the RNA from exonucleases and degradation, and enhancing translation. We became interested in poly(A) tails because they are involved in miRNA-mediated silencing and their lengths seem to be regulated by stress. Using new methods for genome-wide profiling of tail lengths for individual transcripts in C. elegans as well as human cells, we have uncovered previously unrecognized complexities in poly(A) tail length control and are studying the relationship between tail size, proteins that bind poly(A) tails, and the regulation of gene expression.
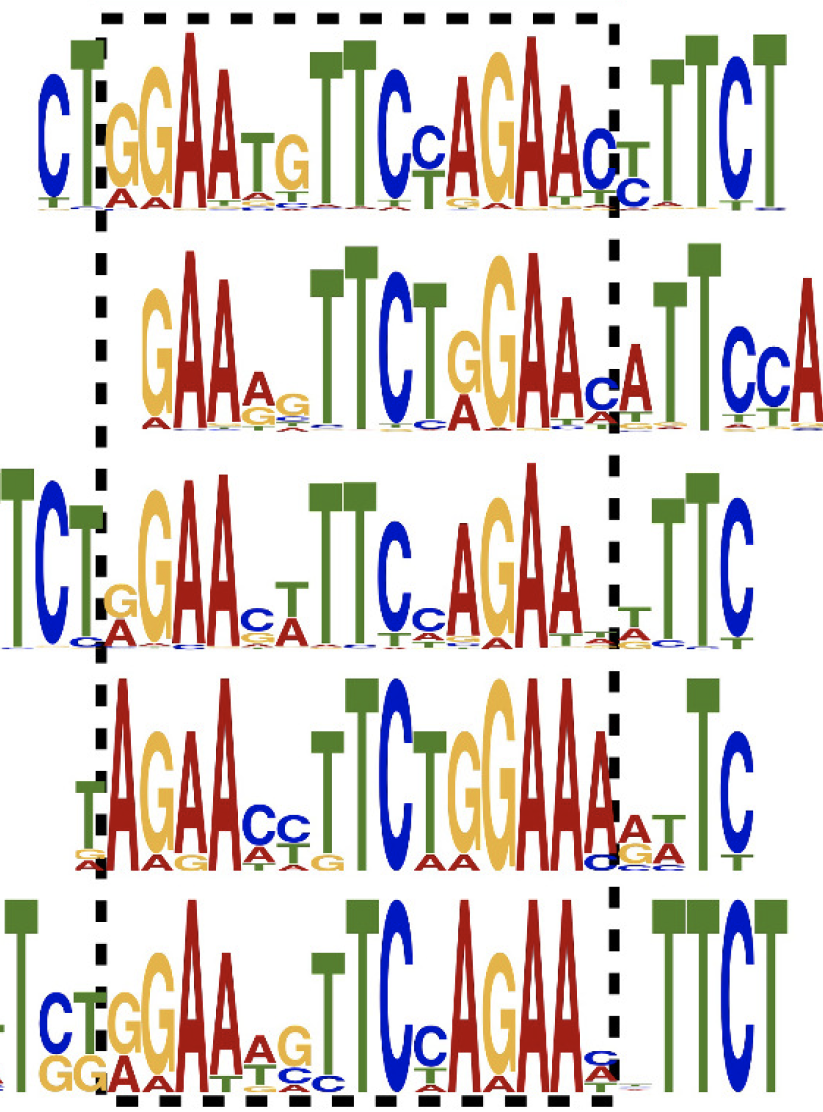
The observation that specific transposon RNAs are induced by heat shock in C. elegans led us to discover that many transposon-derived sequences in this organism harbor transcription factor binding motifs. In one example, we found that Helitron transposons contain a motif important for transcription during heat shock and their proliferation within the genome has brought new genes under the control of the heat shock response. We are curious to explore the sequence information within transposons and ask how these elements might contribute to the regulation of host gene expression.